|
hedgehog signaling pathway
The Hedgehog signaling pathway is a signaling pathway that transmits information to embryonic cells required for proper cell differentiation. Different parts of the embryo have different concentrations of hedgehog signaling proteins. The pathway also has roles in the adult. Diseases associated with the malfunction of this pathway include basal cell carcinoma.
The Hedgehog signaling pathway is one of the key regulators of animal development and is present in all bilaterians. The pathway takes its name from its polypeptide ligand, an intracellular signaling molecule called Hedgehog (Hh) found in fruit flies of the genus Drosophila; fruit fly larva lacking the Hh gene are said to resemble hedgehogs. Hh is one of Drosophila's segment polarity gene products, involved in establishing the basis of the fly body plan. Larvae without Hh are short and spiny, resembling the hedgehog animal. The molecule remains important during later stages of embryogenesis and metamorphosis.
Mammals have three Hedgehog homologues, Desert (DHH), Indian (IHH), and Sonic (SHH), of which Sonic is the best studied. The pathway is equally important during vertebrate embryonic development and is therefore of interest in evolutionary developmental biology. In knockout mice lacking components of the pathway, the brain, skeleton, musculature, gastrointestinal tract and lungs fail to develop correctly. Recent studies point to the role of Hedgehog signaling in regulating adult stem cells involved in maintenance and regeneration of adult tissues. The pathway has also been implicated in the development of some cancers. Drugs that specifically target Hedgehog signaling to fight this disease are being actively developed by a number of pharmaceutical companies. (W)
|
|
hemolysin
Hemolysins or haemolysins are lipids and proteins that cause lysis of red blood cells by disrupting the cell membrane. Although the lytic activity of some microbe-derived hemolysins on red blood cells may be of great importance for nutrient acquisition, many hemolysins produced by pathogens do not cause significant destruction of red blood cells during infection. However, hemolysins are often capable of lysing red blood cells in vitro.
While most hemolysins are protein compounds, some are lipid biosurfactants. (W)
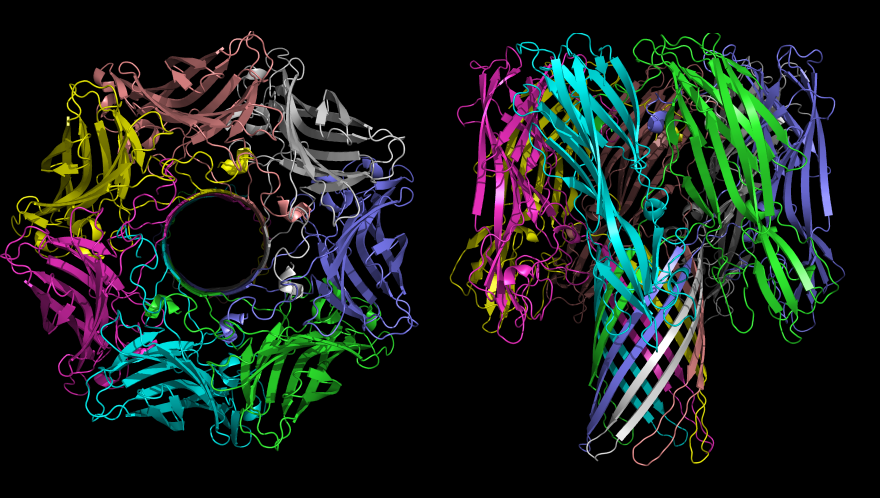
Representation of Alpha Haemloysin from Staph Aureus, created from PDB file 7AHL, using PyMol. Created by me for wikipedia use. |
|
The process of hemolysis in blood cells.
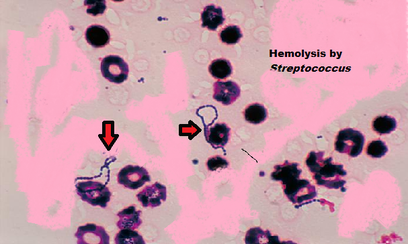
Hemolysis by Streptococcus seen on a plate. |
|
|
|
hemolysis
Hemolysis or haemolysis (also known by several other names, is the rupturing (lysis) of red blood cells (erythrocytes) and the release of their contents (cytoplasm) into surrounding fluid (e.g. blood plasma). Hemolysis may occur in vivo or in vitro (inside or outside the body).
One cause of hemolysis is the action of hemolysins, toxins that are produced by certain pathogenic bacteria or fungi. Another cause is intense physical exercise. Hemolysins damage the red blood cell's cytoplasmic membrane, causing lysis and eventually cell death. (W)

These tubes illustrate different grades of hemolysis, as assessed visually and by our chemistry analyzer, which provides a quantification of the amount of hemoglobin in the sample as a hemolysis index. The hemolysis index (HI) value was determined on the AU5800 analyzer (Beckman Coulter, Brea, CA). The relationship between HI value and concentration of free hemoglobin on the Beckman Coulter analyzers was classified as follows: HI 0: <0,5 g/L, HI 1: 0,5–0,99 g/L, HI 2: 1–1,99 g/L, HI 3: 2–2,99 g/L, HI 4: 3–4,99 g/L |
|

Hemolysis from streptococcus. Examples of the blood culture patterns created by (from left) alpha-, beta- and gamma-hemolytic streptococci. |
|
|
|
heterochromatin
Heterochromatin is a tightly packed form of DNA or condensed DNA, which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed, however according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin.
Constitutive heterochromatin can affect the genes near itself (e.g. position-effect variegation). It is usually repetitive and forms structural functions such as centromeres or telomeres, in addition to acting as an attractor for other gene-expression or repression signals.
Facultative heterochromatin is the result of genes that are silenced through a mechanism such as histone deacetylation or Piwi-interacting RNA (piRNA) through RNAi. It is not repetitive and shares the compact structure of constitutive heterochromatin. However, under specific developmental or environmental signaling cues, it can lose its condensed structure and become transcriptionally active.
Heterochromatin has been associated with the di- and tri-methylation of H3K9 in certain portions of the genome. H3K9me3-related methyltransferases appear to have a pivotal role in modifying heterochromatin during lineage commitment at the onset of organogenesis and in maintaining lineage fidelity.
Note that the informal diagram shown here may be in error as to the location of heterochromatin. An inactivated X-chromosome (a.k.a. Barr body) migrates to the nuclear membrane alone, leaving the active X and other chromosomes within the nucleoplasm (away from the membrane in general). Other heterochromatin appear as particles separate from the membrane, "Heterochromatin appears as small, darkly staining, irregular particles scattered throughout the nucleus ...".(7) (W)
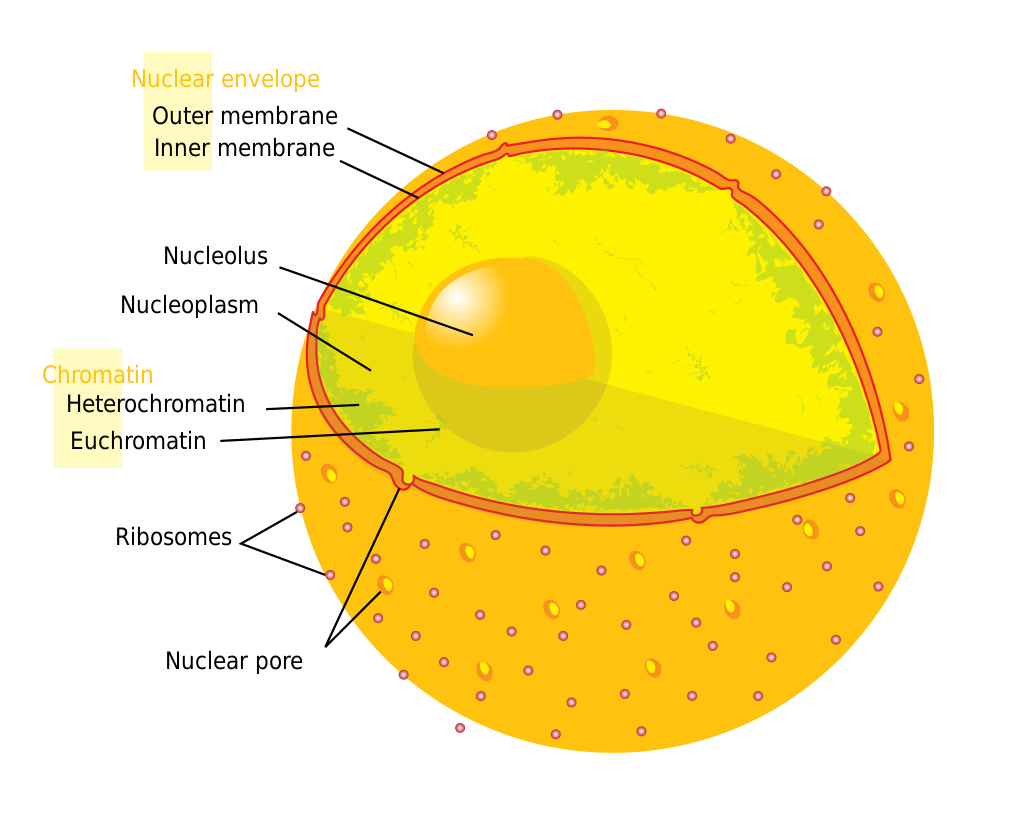
The nucleus of a human cell showing the location of heterochromatin. |
|
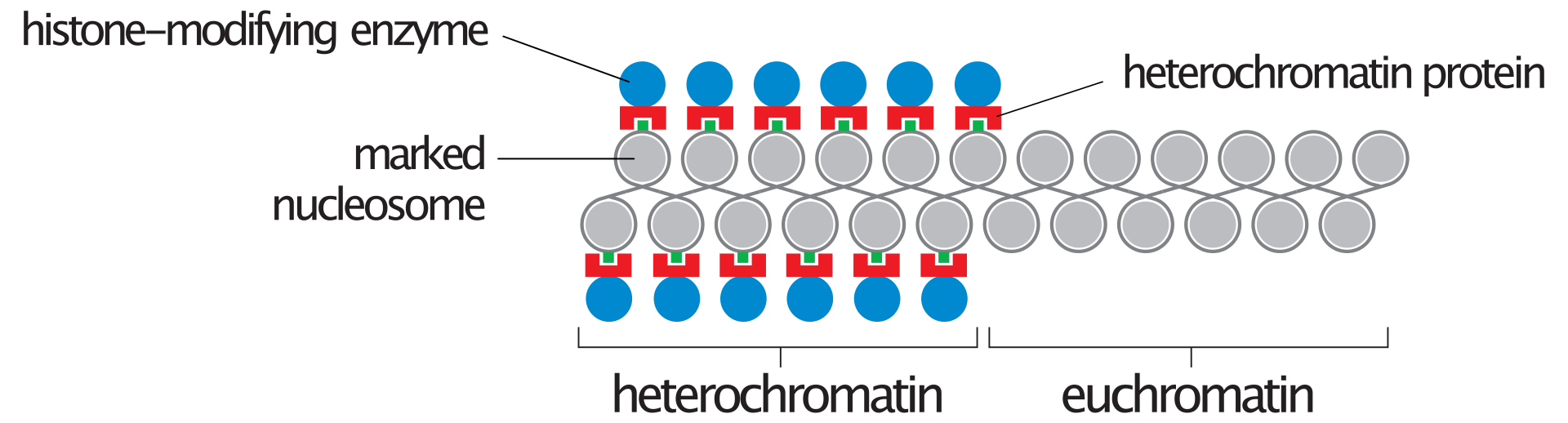
Heterochromatin vs. euchromatin.
Heterochromatin contains specialized proteins (red) that bind to histone H3 or H4 subunits that have been marked by a specific modification (green). The enzyme that performs this modification is also present in heterochromatin (blue), ensuring that the modification is maintained.
|
|
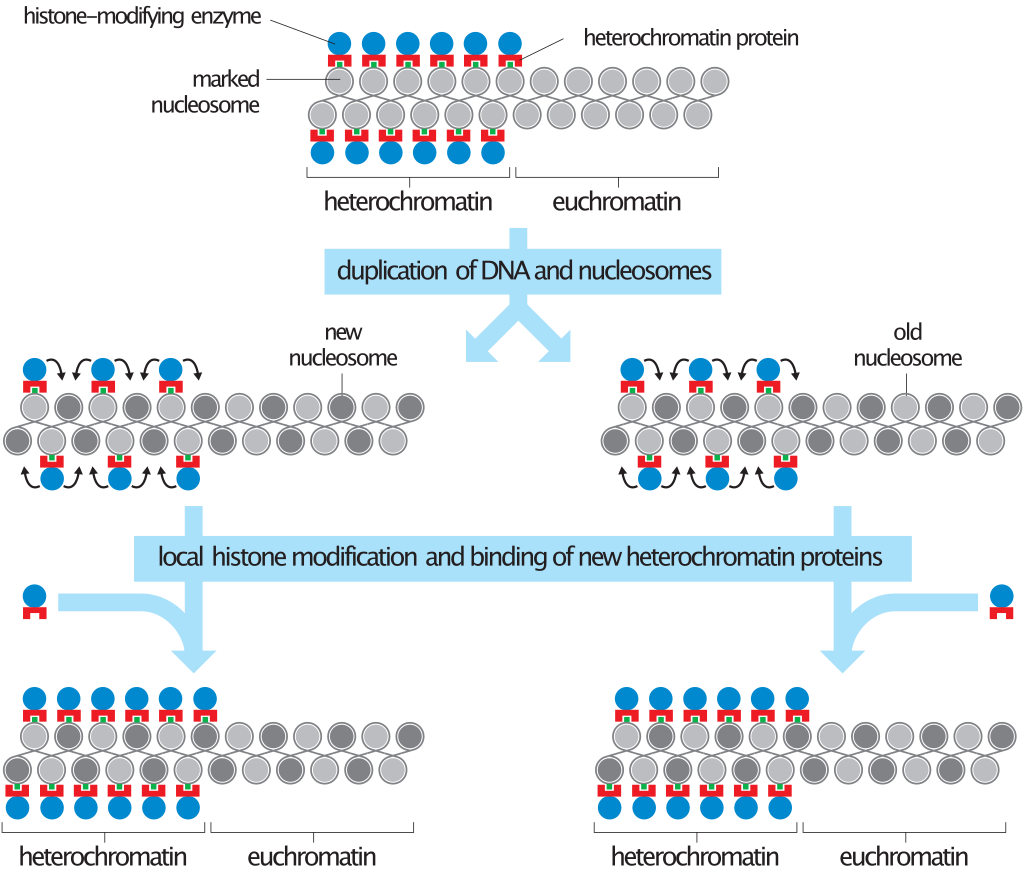
General model for duplication of heterochromatin during cell division.
Heterochromatin contains specialized proteins (red) that bind to histone H3 or H4 subunits that have been marked by a specific modification (green). The enzyme that performs this modification is also present in heterochromatin (blue), ensuring that the modification is maintained. When the chromosome is duplicated, the marked histone H3–H4 tetramers of the parent chromosome are distributed randomly to the two daughter strands, resulting in a mixture of old (light grey) and new (dark grey) nucleosomes. In heterochromatin, new nucleosomes are rapidly marked by the histone-modifying enzymes bound to old nucleosomes. This provides new binding sites for heterochromatin proteins. These proteins (such as the Sir complex or HP1) also have the ability to bind to each other, further promoting the assembly of a protein polymer along the chromosome. It can also be seen in this diagram that the boundary between heterochromatin and euchromatin is not rigidly fixed, because small local changes in the extent of histone modification could cause shifts in its position. |
|
|
|
HindIII
HindIII (pronounced "Hin D Three") is a type II site-specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis.
The cleavage of this sequence between the AA's results in 5' overhangs on the DNA called sticky ends:
5'-A |A G C T T-3'
3'-T T C G A| A-5'
Restriction endonucleases are used as defense mechanisms in prokaryotic organisms in the restriction modification system. Their primary function is to protect the host genome against invasion by foreign DNA, primarily bacteriophage DNA. There is also evidence that suggests the restriction enzymes may act alongside modification enzymes as selfish elements, or may be involved in genetic recombination and transposition. (W)
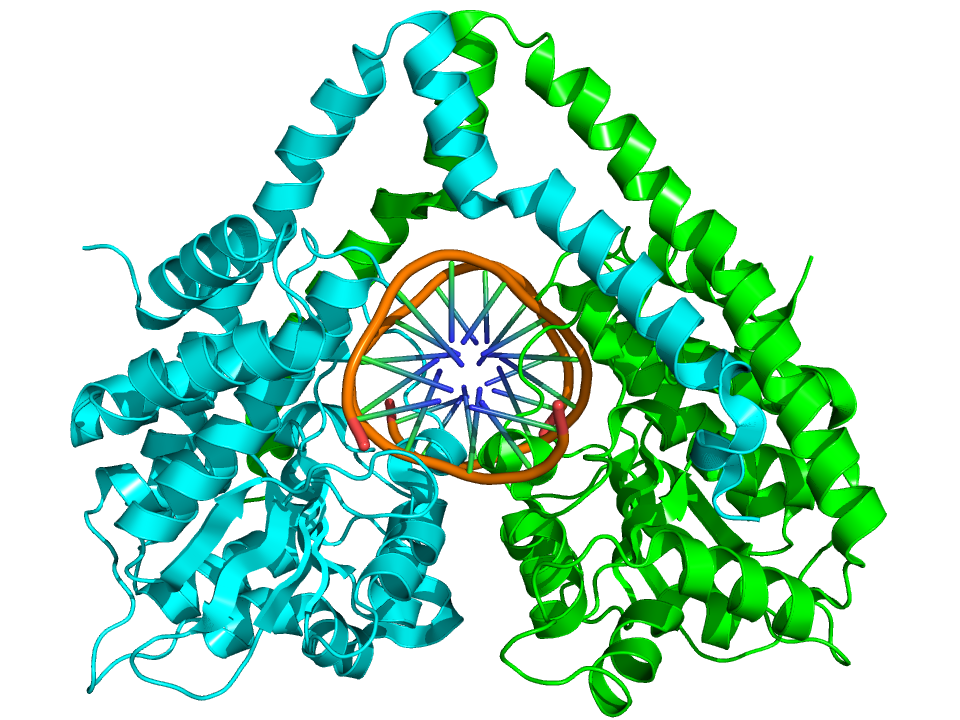
Crystallographic structure of the HindIII restriction endonuclease dimer (cyan and green) complexed with double helical DNA (brown) based on the PDB: 2E52 coordinates. |
|
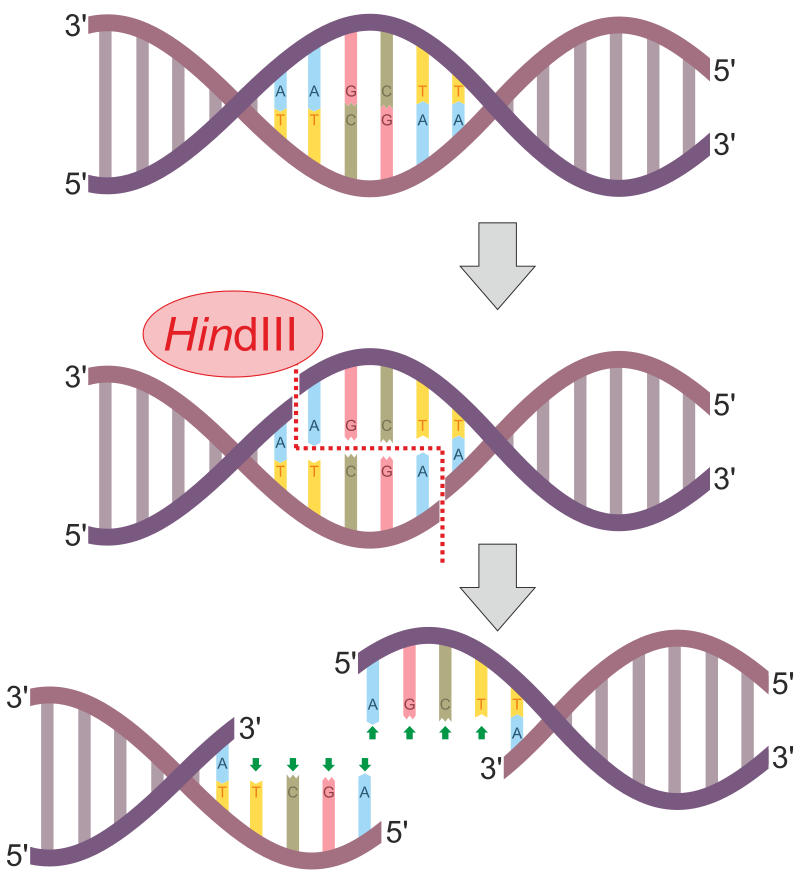
HindIII restrictions process results in formation of overhanging palindromic sticky ends. |
|
BglII catalytic site, showing the coordination of Asp 84 and Mg2+ with water. |
|
|
|
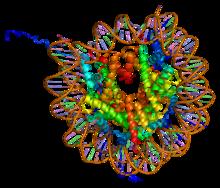
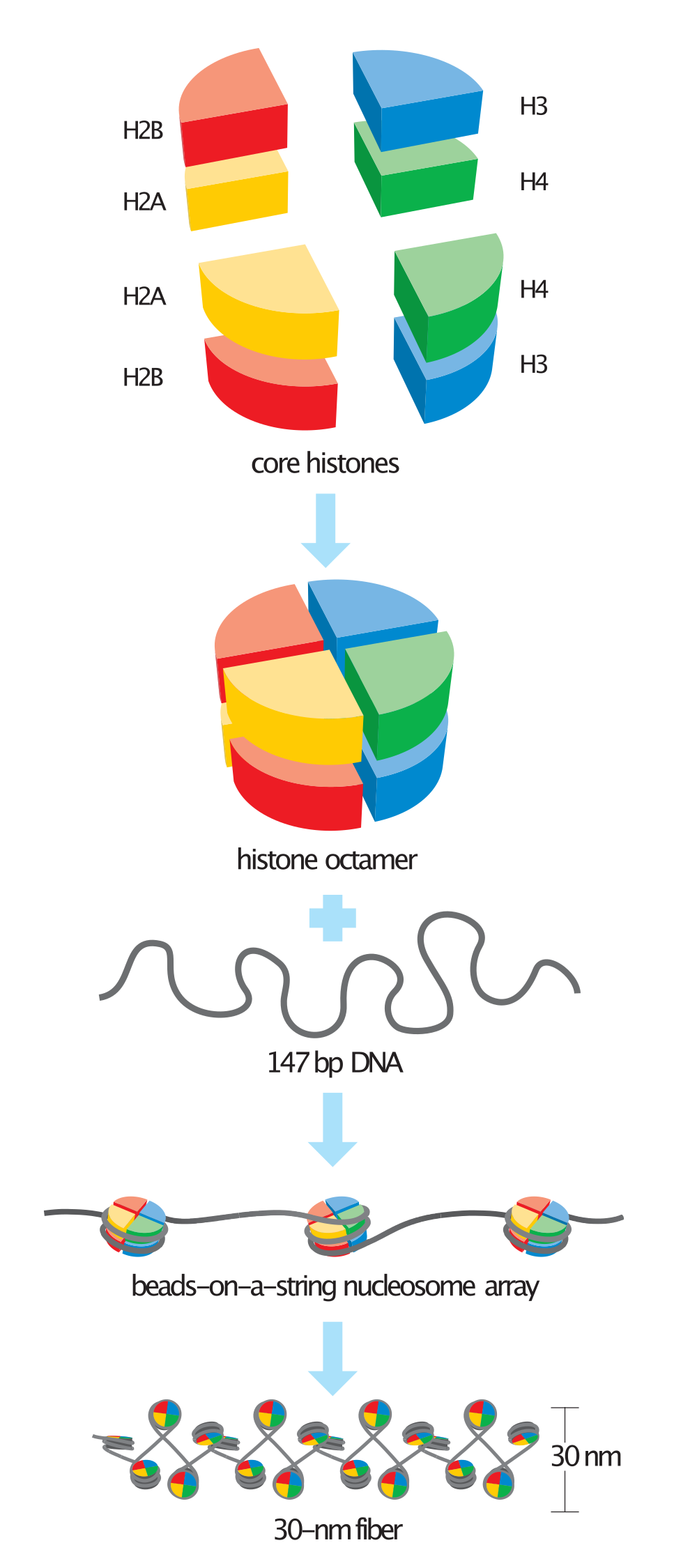
histone
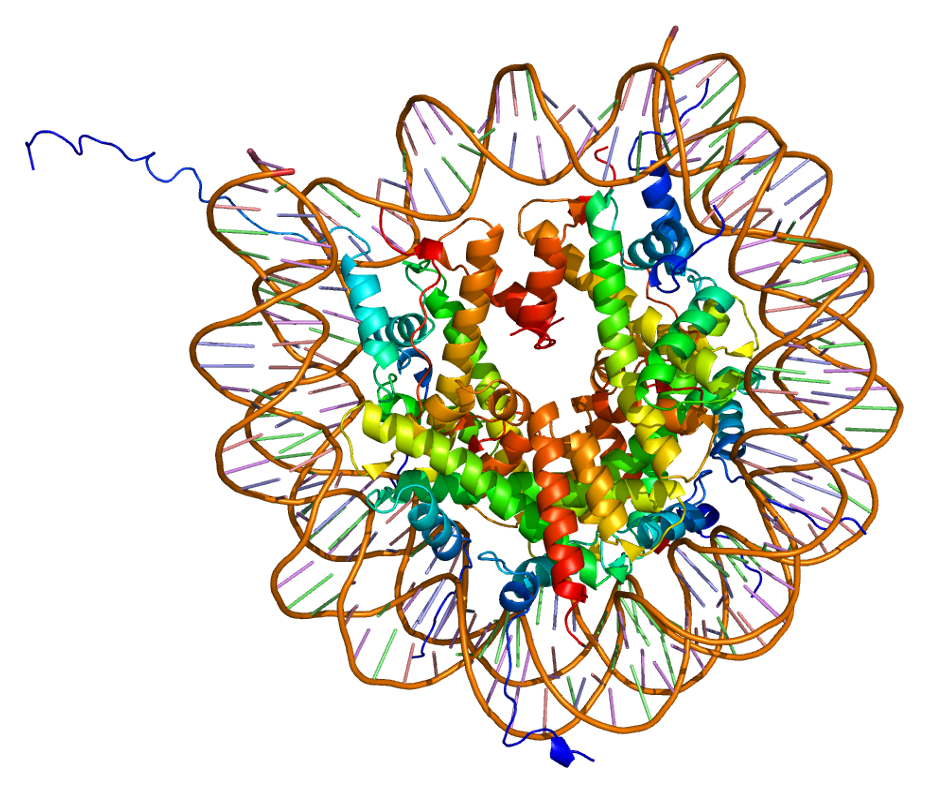
In biology, histones are highly basic proteins found in eukaryotic cell nuclei that pack and order the DNA into structural units called nucleosomes. Histones are abundant in lysine and arginine. Histones are the chief protein components of chromatin, acting as spools around which DNA winds, and playing a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long (a length to width ratio of more than 10 million to 1 in human DNA). For example, each human diploid cell (containing 23 pairs of chromosomes) has about 1.8 meters of DNA; wound on the histones, the diploid cell has about 90 micrometers (0.09 mm) of chromatin. When the diploid cells are duplicated and condensed during mitosis, the result is about 120 micrometers of chromosomes
Classes and histone variants
Five major families of histones exist: H1/H5, H2A, H2B, H3, and H4. Histones H2A, H2B, H3 and H4 are known as the core histones, while histones H1/H5 are known as the linker histones.
(W)
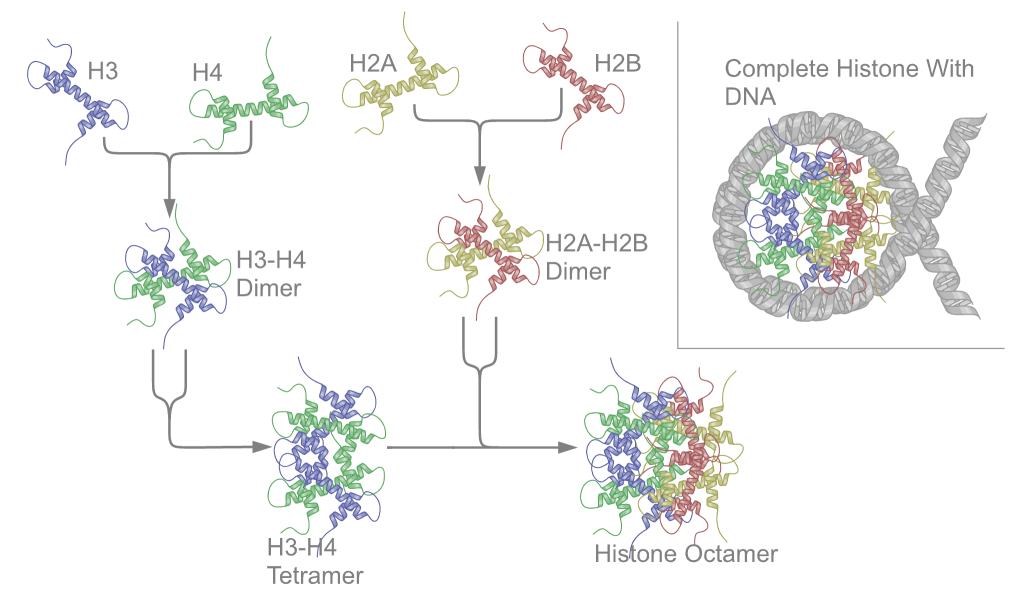
Schematic representation of the assembly of the core histones into the nucleosome. |
|
Structure of the H2AFJ protein. Based on PyMOL rendering of PDB 1aoi. |
|
linker histone H1 and H5 family. PDB rendering of HIST1H1B based on 1ghc.
|
|
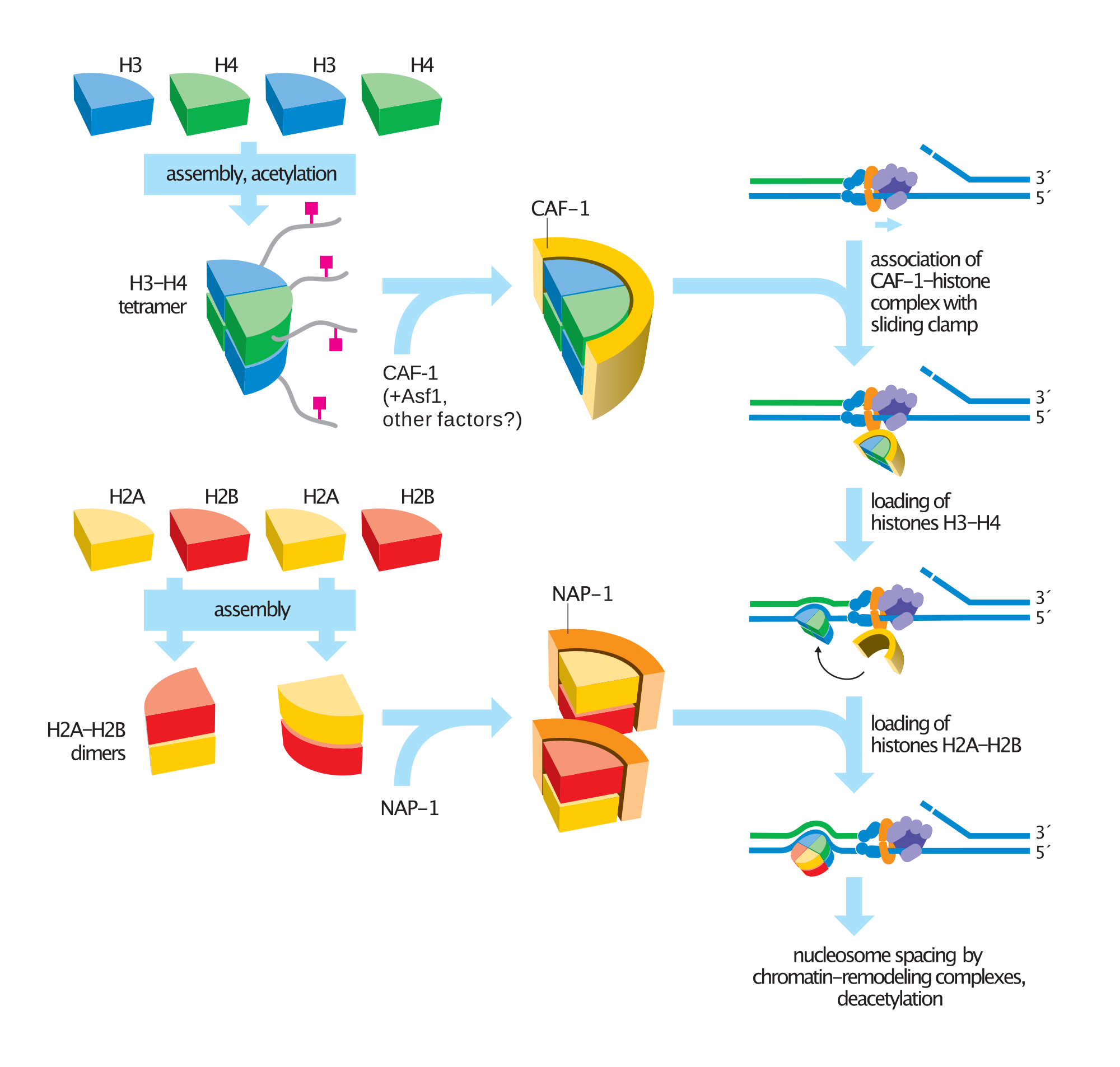
Steps in nucleosome assembly. |
|
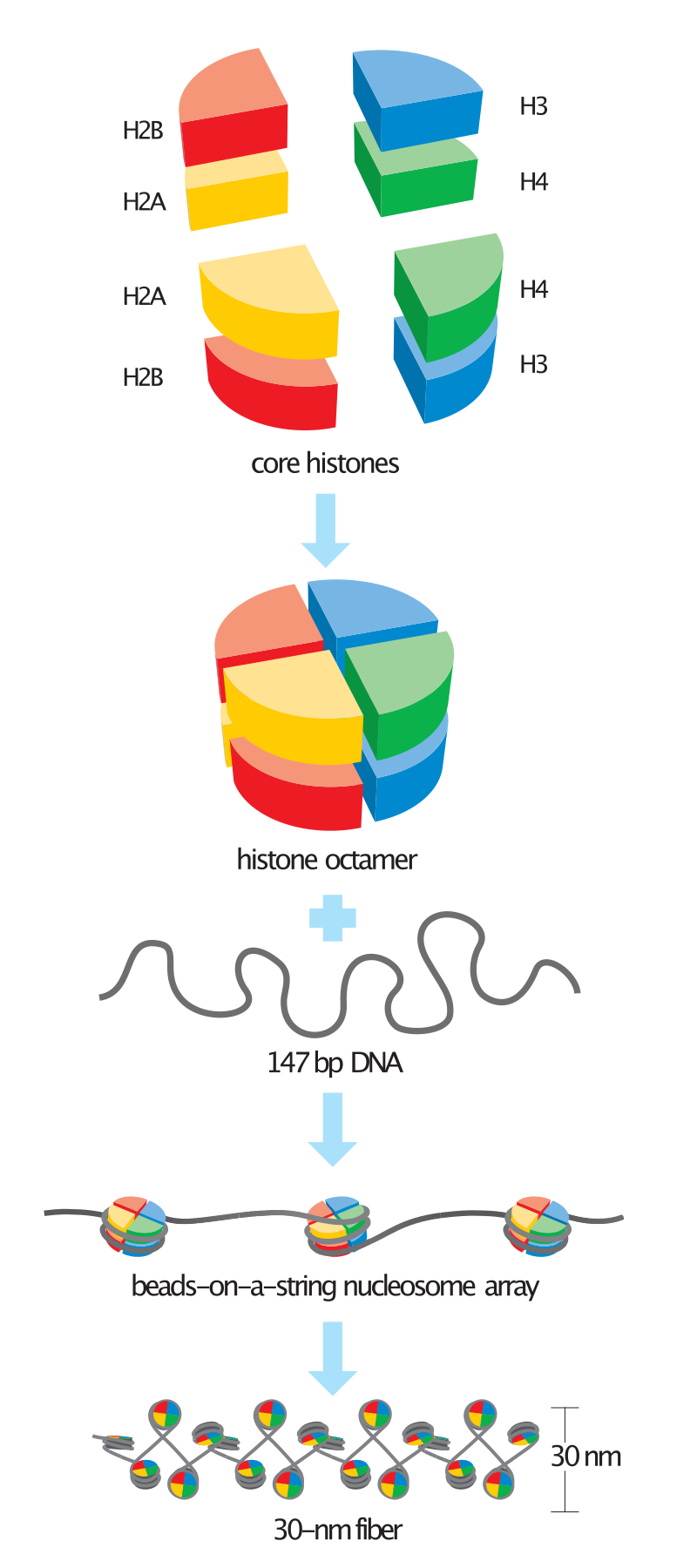
Basic units of chromatin structure. |
|
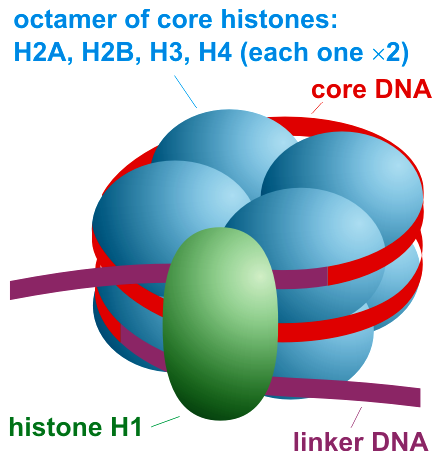
Scheme of nucleosome organization. |
|
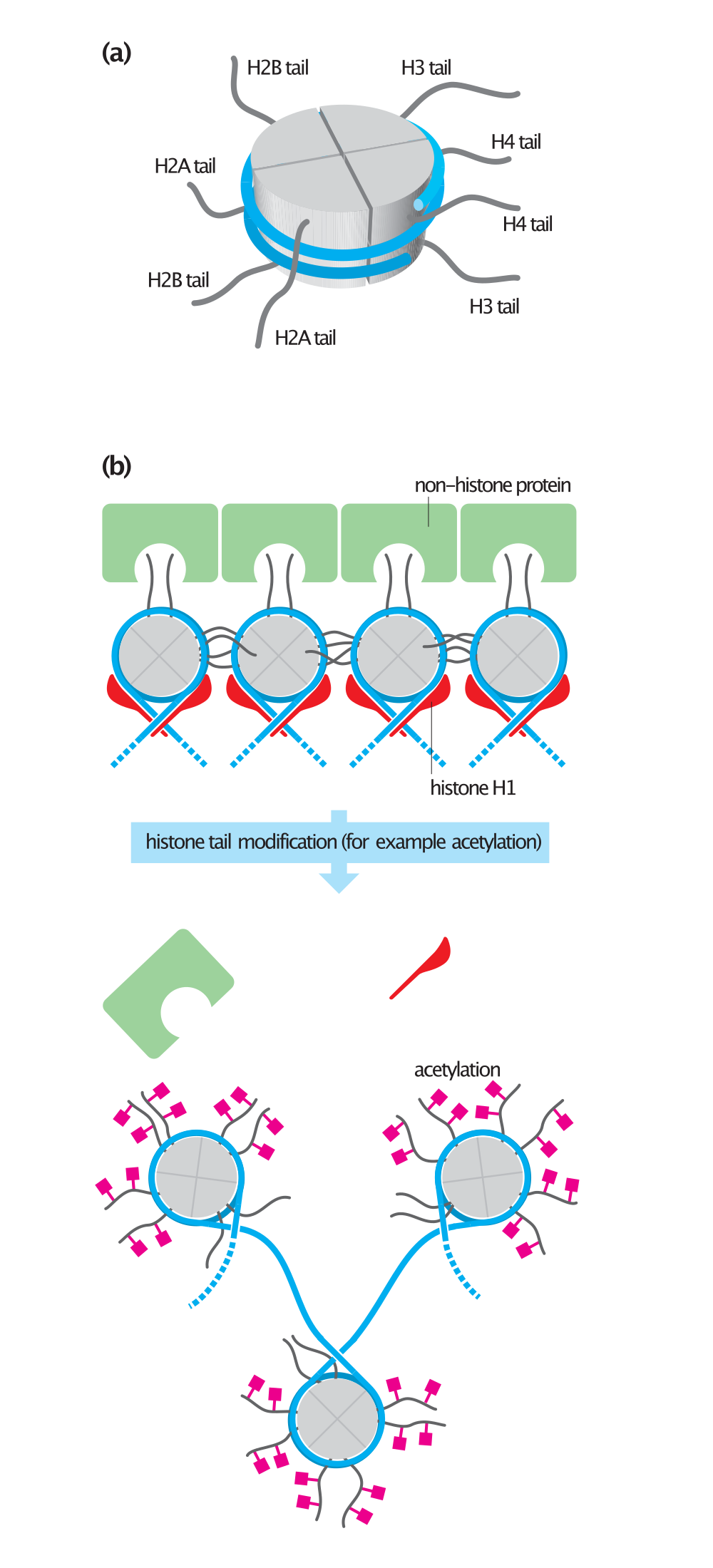
Histone tails and their function in chromatin formation. |
|
Functions of histone modifications (L)
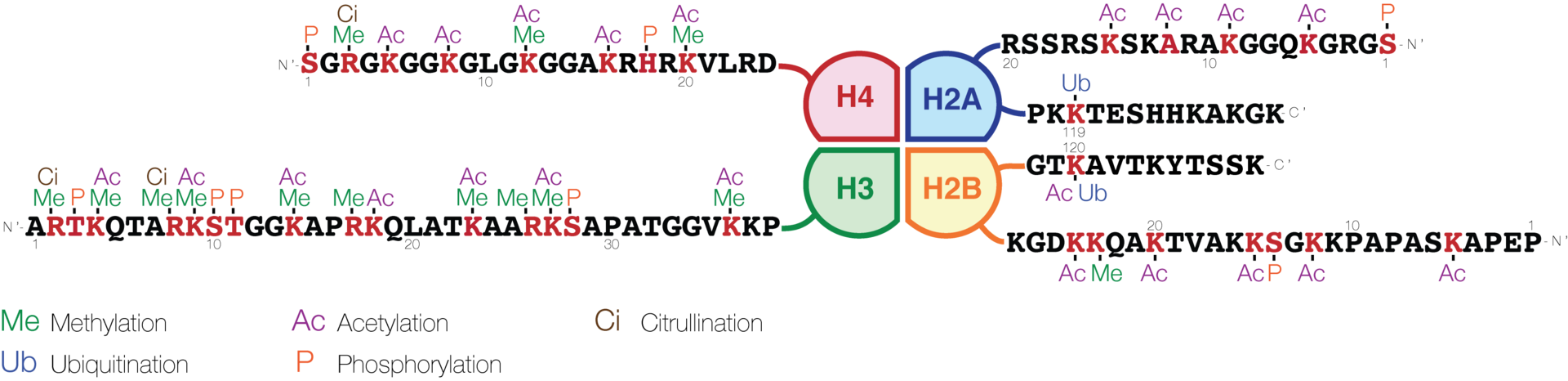
Schematic representation of histone modifications. Based on Rodriguez-Paredes and Esteller, Nature, 2011
A huge catalogue of histone modifications have been described, but a functional understanding of most is still lacking. Collectively, it is thought that histone modifications may underlie a histone code, whereby combinations of histone modifications have specific meanings. However, most functional data concerns individual prominent histone modifications that are biochemically amenable to detailed study. |
|
|
histone: H1/H5, H2A, H2B, H3, and H4
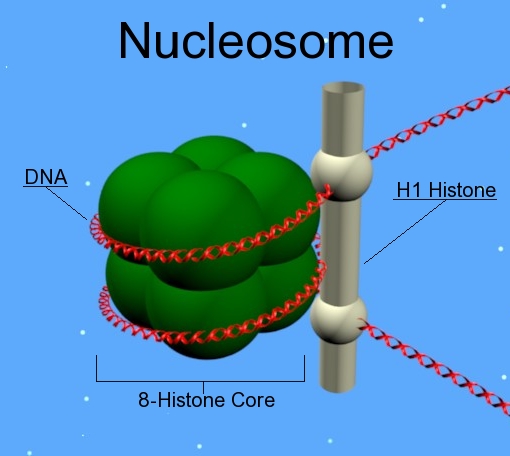
A diagram showing where H1 can be found in the nucleosome.
Histone H1 is one of the five main histone protein families which are components of chromatin in eukaryotic cells. Though highly conserved, it is nevertheless the most variable histone in sequence across species. (W)
|
|
Structure of the H2AFJ protein.
Histone H2A is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells.highly conserved, it is nevertheless the most variable histone in sequence across species. (W)
|
|
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure. Histone proteins are highly post-translationally modified however Histone H3 is the most extensively modified of the five histones. The term "Histone H3" alone is purposely ambiguous in that it does not distinguish between sequence variants or modification state. Histone H3 is an important protein in the emerging field of epigenetics, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. (W)
|
|

Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on a string' organization. Histone proteins are highly post-translationally modified. Covalently bonded modifications include acetylation and methylation of the N-terminal tails. These modifications may alter expression of genes located on DNA associated with its parent histone octamer. Histone H4 is an important protein in the structure and function of chromatin, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. (W) |
|
|
|
histone methylation
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes. (W)
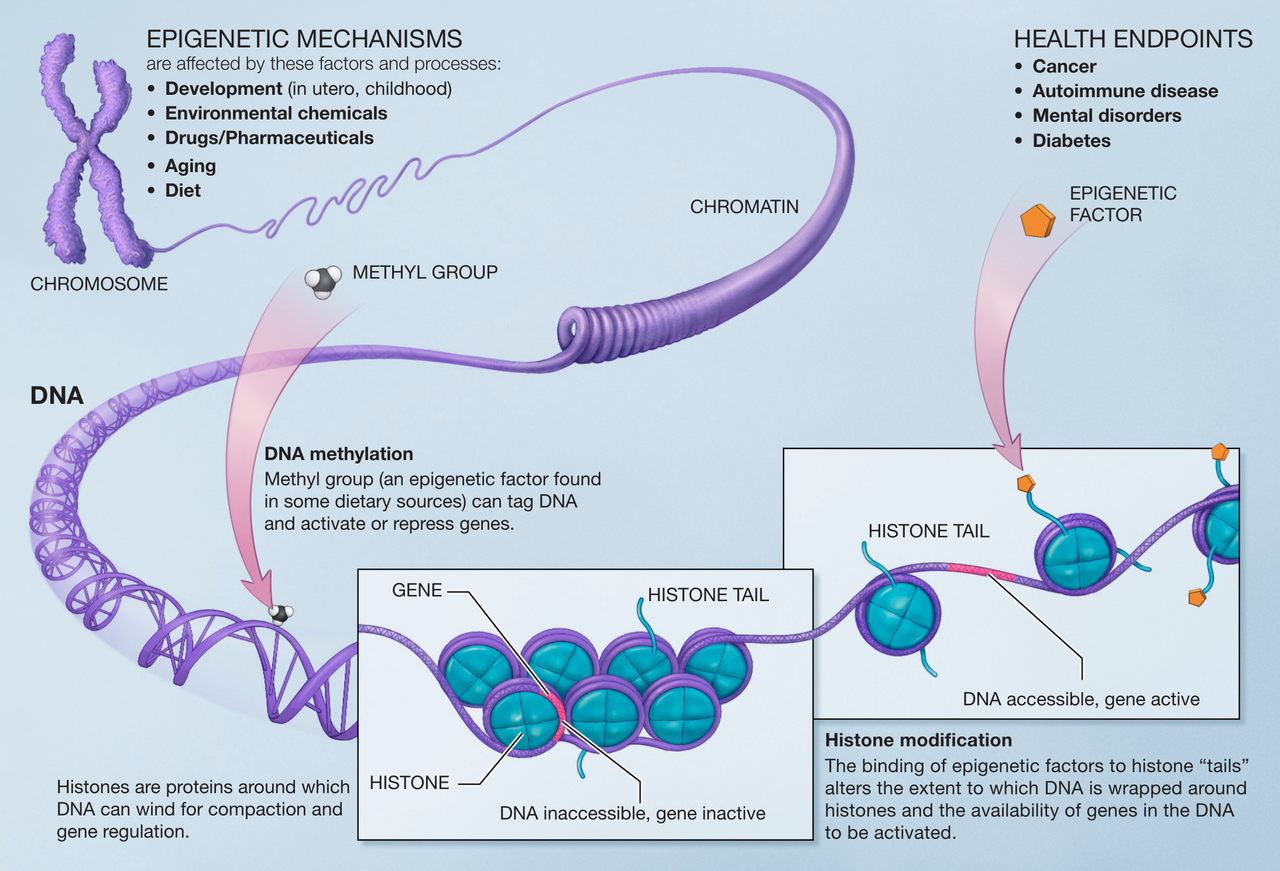
Epigenetic mechanisms.
Epigenetic mechanisms are affected by several factors and processes including development in utero and in childhood, environmental chemicals, drugs and pharmaceuticals, aging, and diet. DNA methylation is what occurs when methyl groups, an epigenetic factor found in some dietary sources, can tag DNA and activate or repress genes. Histones are proteins around which DNA can wind for compaction and gene regulation. Histone modification occurs when the binding of epigenetic factors to histone “tails” alters the extent to which DNA is wrapped around histones and the availability of genes in the DNA to be activated. All of these factors and processes can have an effect on people’s health and influence their health possibly resulting in cancer, autoimmune disease, mental disorders, or diabetes among other illnesses. National Institutes of Health |
|
|
|
homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may be also RNA in viruses). It is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses. (W)
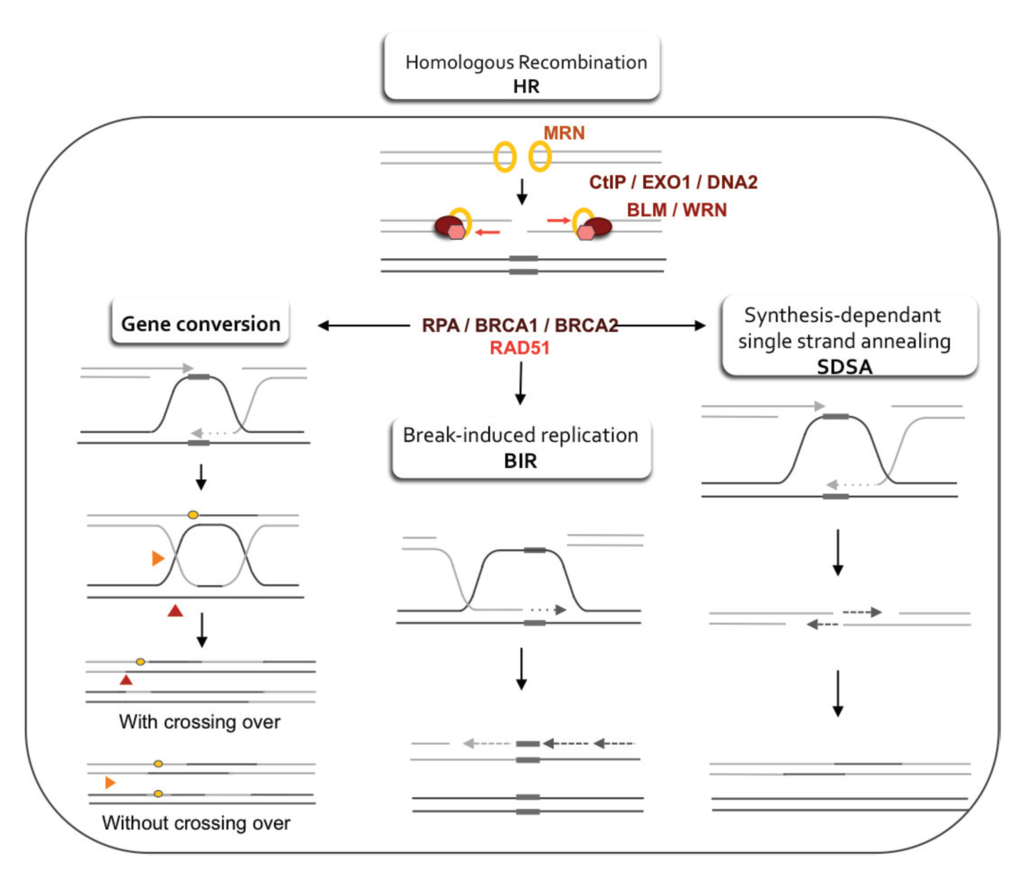
Figure 4. Double-strand break repair models that act via homologous recombination.
Left panel: Gene conversion. After resection, the single-stranded 3' tail invades a homologous, intact double-stranded DNA, forming a D-loop (displacement loop). This process tolerates a limited number of imperfect sequence homologies, thus creating heteroduplex intermediates bearing mismatches (yellow circles). The invading 3'-end primes DNA synthesis, which then fills in the gaps. The cruciform junctions (Holliday junctions, HJ) migrate. Resolution (or dissolution) of HJs occurs in two different orientations (orange or red triangles), resulting in gene conversion either with or without crossing over. Middle panel: Break-induced replication (BIR). The initiation is similar to that of the previous models, but the synthesis continues over longer distances on the chromosome arms, even reaching the end of the chromosome. Here, there is neither resolution of the HR nor crossover. Right panel: Synthesis-dependent strand annealing (SDSA). Initiation is similar to that of the previous model, but the invading strand de-hybridizes and re-anneals at the other end of the injured molecule; no HJ is formed. (W) |
|
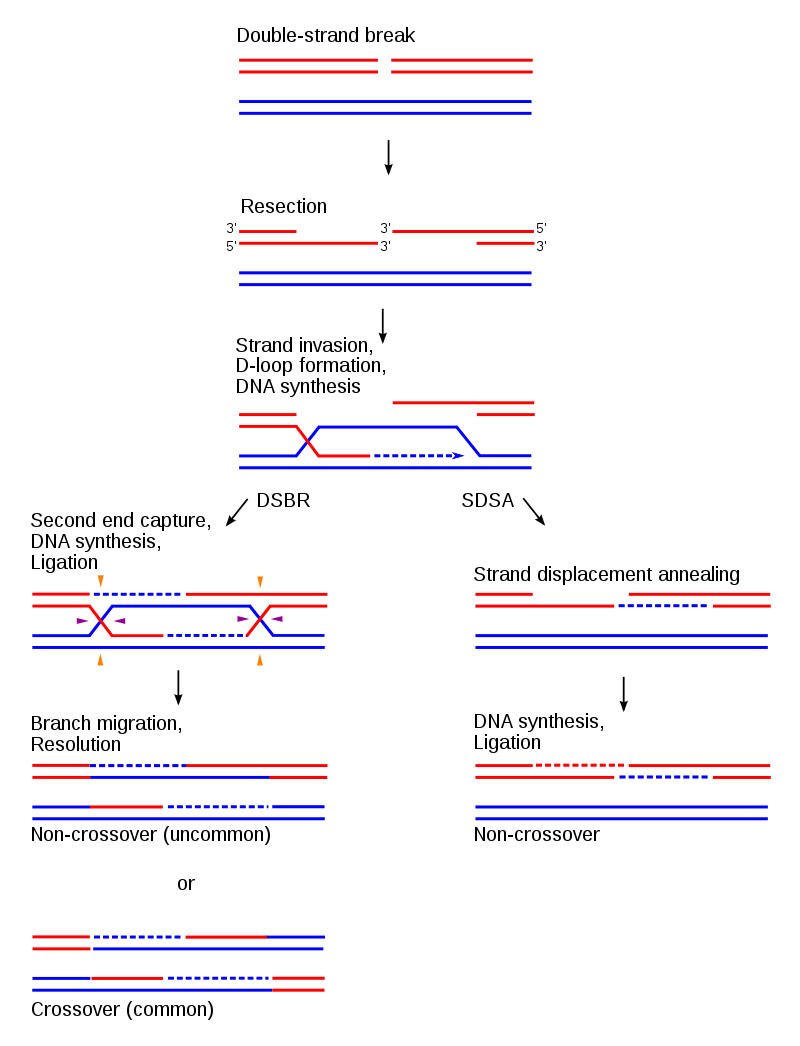
Figure 5. The DSBR and SDSA pathways follow the same initial steps, but diverge thereafter. The DSBR pathway most often results in chromosomal crossover (bottom left), while SDSA always ends with non-crossover products (bottom right). |
|
|
|
homology directed repair
Homology directed repair (HDR) is a mechanism in cells to repair double-strand DNA lesions. The most common form of HDR is homologous recombination. The HDR mechanism can only be used by the cell when there is a homologous piece of DNA present in the nucleus, mostly in G2 and S phase of the cell cycle. Other examples of homology-directed repair include single-strand annealing and breakage-induced replication. When the homologous DNA is absent, another process called non-homologous end joining (NHEJ) takes place instead. (W)

Double-strand break repair models that act via homologous recombination.
Left panel: Gene conversion. After resection, the single-stranded 3' tail invades a homologous, intact double-stranded DNA, forming a D-loop (displacement loop). This process tolerates a limited number of imperfect sequence homologies, thus creating heteroduplex intermediates bearing mismatches (yellow circles). The invading 3'-end primes DNA synthesis, which then fills in the gaps. The cruciform junctions (Holliday junctions, HJ) migrate. Resolution (or dissolution) of HJs occurs in two different orientations (orange or red triangles), resulting in gene conversion either with or without crossing over. Middle panel: Break-induced replication (BIR). The initiation is similar to that of the previous models, but the synthesis continues over longer distances on the chromosome arms, even reaching the end of the chromosome. Here, there is neither resolution of the HR nor crossover. Right panel: Synthesis-dependent strand annealing (SDSA). Initiation is similar to that of the previous model, but the invading strand de-hybridizes and re-anneals at the other end of the injured molecule; no HJ is formed. |
|
|
|
horizontal gene transfer
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between unicellular and/or multicellular organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms.
Horizontal gene transfer is the primary mechanism for the spread of antibiotic resistance in bacteria, and plays an important role in the evolution of bacteria that can degrade novel compounds such as human-created pesticides and in the evolution, maintenance, and transmission of virulence. It often involves temperate bacteriophages and plasmids. Genes responsible for antibiotic resistance in one species of bacteria can be transferred to another species of bacteria through various mechanisms of HGT such as transformation, transduction and conjugation, subsequently arming the antibiotic resistant genes' recipient against antibiotics. The rapid spread of antibiotic resistance genes in this manner is becoming medically challenging to deal with. Ecological factors may also play a role in the LGT of antibiotic resistant genes. It is also postulated that HGT promotes the maintenance of a universal life biochemistry and, subsequently, the universality of the genetic code.
Most thinking in genetics has focused upon vertical transfer, but the importance of horizontal gene transfer among single-cell organisms is beginning to be acknowledged.
Gene delivery can be seen as an artificial horizontal gene transfer, and is a form of genetic engineering. (W)
.png)
Horizontal gene transfer. |
|
|
|
human mitochondrial genetics
Human mitochondrial genetics is the study of the genetics of human mitochondrial DNA (the DNA contained in human mitochondria). The human mitochondrial genome is the entirety of hereditary information contained in human mitochondria. Mitochondria are small structures in cells that generate energy for the cell to use, and are hence referred to as the "powerhouses" of the cell.
Mitochondrial DNA (mtDNA) is not transmitted through nuclear DNA (nDNA). In humans, as in most multicellular organisms, mitochondrial DNA is inherited only from the mother's ovum. There are theories, however, that paternal mtDNA transmission in humans can occur under certain circumstances.
Mitochondrial inheritance is therefore non-Mendelian, as Mendelian inheritance presumes that half the genetic material of a fertilized egg (zygote) derives from each parent.
Eighty percent of mitochondrial DNA codes for mitochondrial RNA, and therefore most mitochondrial DNA mutations lead to functional problems, which may be manifested as muscle disorders (myopathies).
Because they provide 30 molecules of ATP per glucose molecule in contrast to the 2 ATP molecules produced by glycolysis, mitochondria are essential to all higher organisms for sustaining life. The mitochondrial diseases are genetic disorders carried in mitochondrial DNA, or nuclear DNA coding for mitochondrial components. Slight problems with any one of the numerous enzymes used by the mitochondria can be devastating to the cell, and in turn, to the organism. (W)
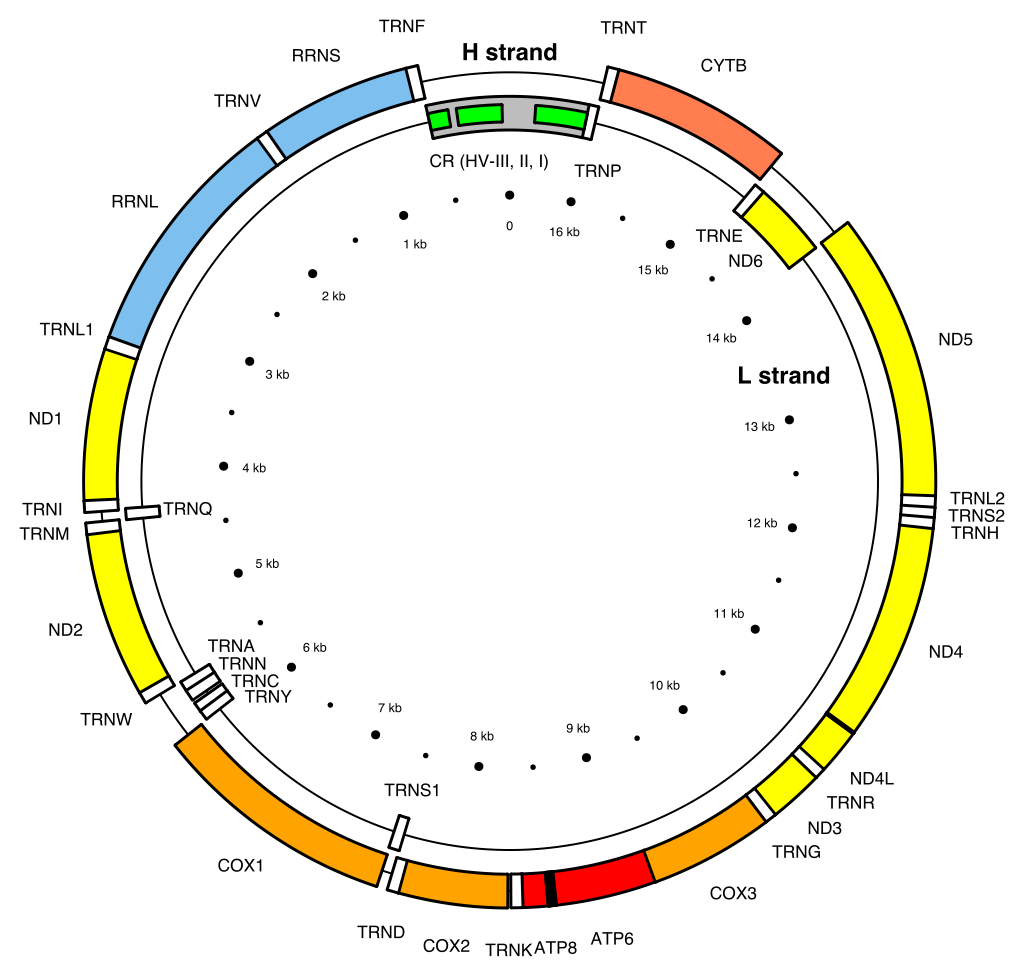
Map of the human mitochondrial DNA genome (16569 bp, NCBI sequence accession NC_012920 — Anderson et al. 1981). The H (heavy, outer circle) and L (light, inner circle) strands are given with their corresponding genes. There are 22 transfer RNA (TRN) genes for the following amino acids: F, V, L1 (codon UUA/G), I, Q, M, W, A, N, C, Y, S1 (UCN), D, K, G, R, H, S2 (AGC/U), L2 (CUN), E, T and P (white boxes). There are 2 ribosomal RNA (RRN) genes: S (small subunit, or 12S) and L (large subunit, or 16S) (blue boxes). There are 13 protein-coding genes: 7 for NADH dehydrogenase subunits (ND, yellow boxes), 3 for cytochrome c oxidase subunits (COX, orange boxes), 2 for ATPase subunits (ATP, red boxes), and one for cytochrome b (CYTB, coral box). Two gene overlaps are indicated (ATP8-ATP6, and ND4L-ND4, black boxes). The control region (CR) is the longest non-coding sequence (grey box). Its three hyper-variable regions are indicated (HV, green boxes). |
|
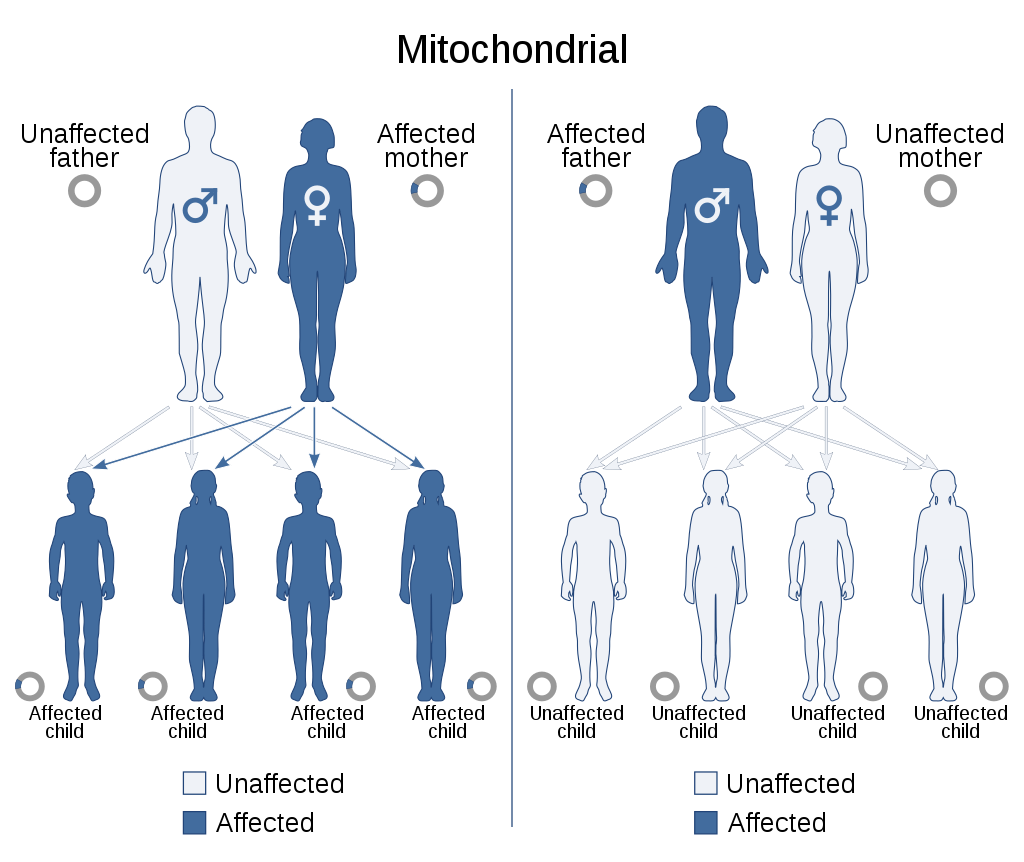
Mitochondrial inheritance patterns. |
|
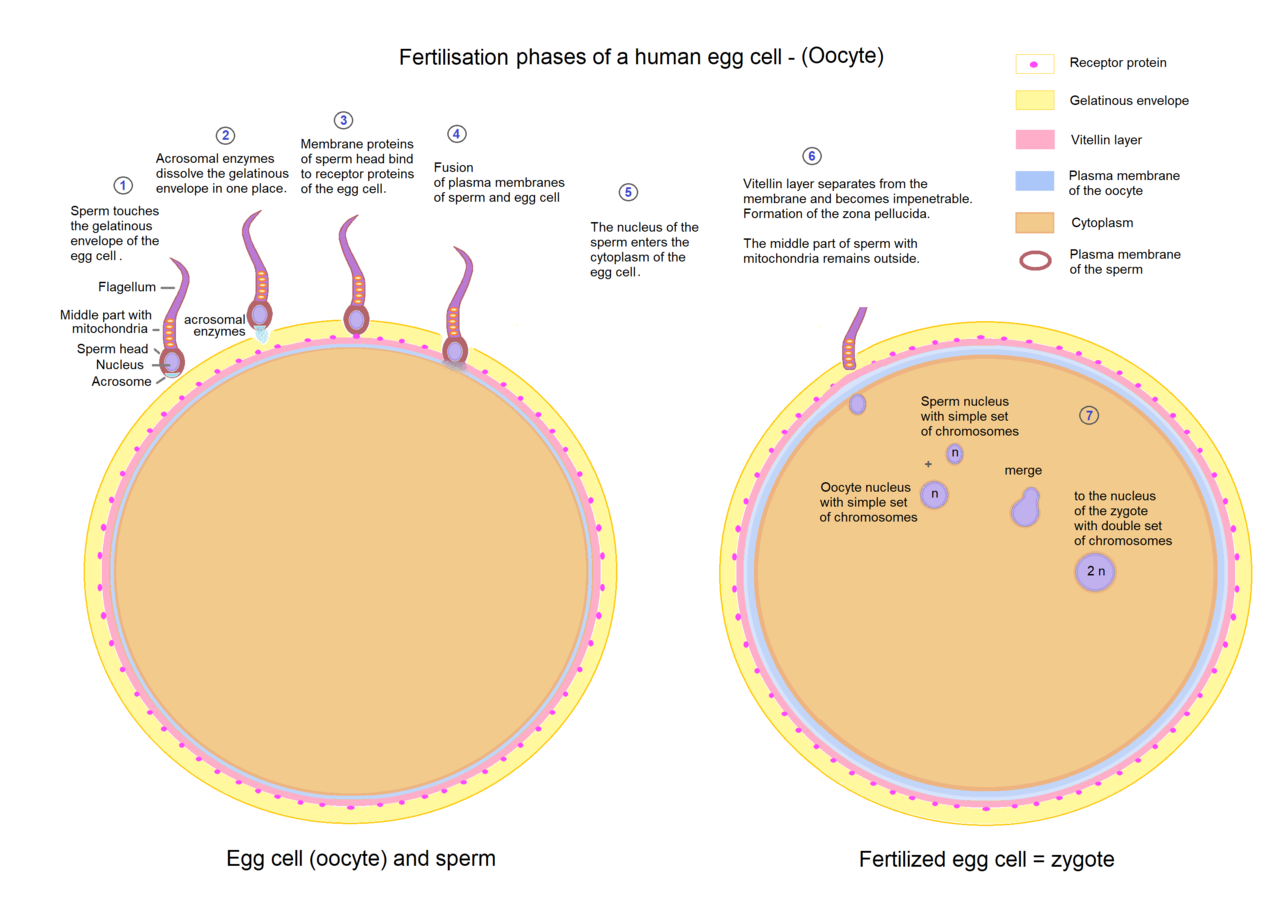
The reason for maternal inheritance in mitochondrial DNA is that when the sperm enters the egg cell, it discards its middle part, which contains its mitochondria, so that only its head with the nucleus penetrates the egg cell. |
|
|
|
huntingtin
Clinical significance
The huntingtin gene, also called the HTT or HD (Huntington disease) gene, is the IT15 ("interesting transcript 15") gene, which codes for a protein called the huntingtin protein. The gene and its product are under heavy investigation as part of Huntington's disease clinical research and the suggested role for huntingtin in long-term memory storage.
It is variable in its structure, as the many polymorphisms of the gene can lead to variable numbers of glutamine residues present in the protein. In its wild-type (normal) form, it contains 6-35 glutamine residues. However, in individuals affected by Huntington's disease (an autosomal dominant genetic disorder), it contains more than 36 glutamine residues (highest reported repeat length is about 250). Its commonly used name is derived from this disease; previously, the IT15 label was commonly used. (W)
|
|
| hybridization → nucleic acid hybridization |
|
hybridization probe
In molecular biology, a hybridization probe is a fragment of DNA or RNA of variable length (usually 100–10000 bases long) which can be radioactively or fluorescently labeled. It can then be used in DNA or RNA samples to detect the presence of nucleotide substances (the RNA target) that are complementary to the sequence in the probe. The probe thereby hybridizes to single-stranded nucleic acid (DNA or RNA) whose base sequence allows probe–target base pairing due to complementarity between the probe and target. The labeled probe is first denatured (by heating or under alkaline conditions such as exposure to sodium hydroxide) into single stranded DNA (ssDNA) and then hybridized to the target ssDNA (Southern blotting) or RNA (northern blotting) immobilized on a membrane or in situ. To detect hybridization of the probe to its target sequence, the probe is tagged (or "labeled") with a molecular marker of either radioactive or (more recently) fluorescent molecules; commonly used markers are 32P (a radioactive isotope of phosphorus incorporated into the phosphodiester bond in the probe DNA) or digoxigenin, which is a non-radioactive, antibody-based marker. DNA sequences or RNA transcripts that have moderate to high sequence similarity to the probe are then detected by visualizing the hybridized probe via autoradiography or other imaging techniques. Normally, either X-ray pictures are taken of the filter, or the filter is placed under UV light. Detection of sequences with moderate or high similarity depends on how stringent the hybridization conditions were applied—high stringency, such as high hybridization temperature and low salt in hybridization buffers, permits only hybridization between nucleic acid sequences that are highly similar, whereas low stringency, such as lower temperature and high salt, allows hybridization when the sequences are less similar. Hybridization probes used in DNA microarrays refer to DNA covalently attached to an inert surface, such as coated glass slides or gene chips, to which a mobile cDNA target is hybridized.
Depending on the method, the probe may be synthesized using the phosphoramidite method, or it can be generated and labeled by PCR amplification or cloning (both are older methods). In order to increase the in vivo stability of the probe RNA is not used. Instead, RNA analogues may be used, in particular morpholino- derivatives. Molecular DNA- or RNA-based probes are now routinely used in screening gene libraries, detecting nucleotide sequences with blotting methods, and in other gene technologies, such as nucleic acid and tissue microarrays. (W)

Fluorescence Microscope and Pipettes.
A technician viewing a blot on a fluorescence microscope while another technician is using a pipette at the Advanced Technology Research Facility (ATRF), Frederick National Laboratory for Cancer Research, National Cancer Institute. (L) |
|
|
|
hydrate
In chemistry, a hydrate is a substance that contains water or its constituent elements. The chemical state of the water varies widely between different classes of hydrates, some of which were so labeled before their chemical structure was understood. (W) |
|
hydrocarbon
In organic chemistry, a hydrocarbon is an organic compound consisting entirely of hydrogen and carbon. 620 Hydrocarbons are examples of group 14 hydrides. Hydrocarbons from which one hydrogen atom has been removed are functional groups called hydrocarbyls. Hydrocarbons are generally colorless and hydrophobic with only weak odors. Because of their diverse molecular structures, it is difficult to generalize further. (W)
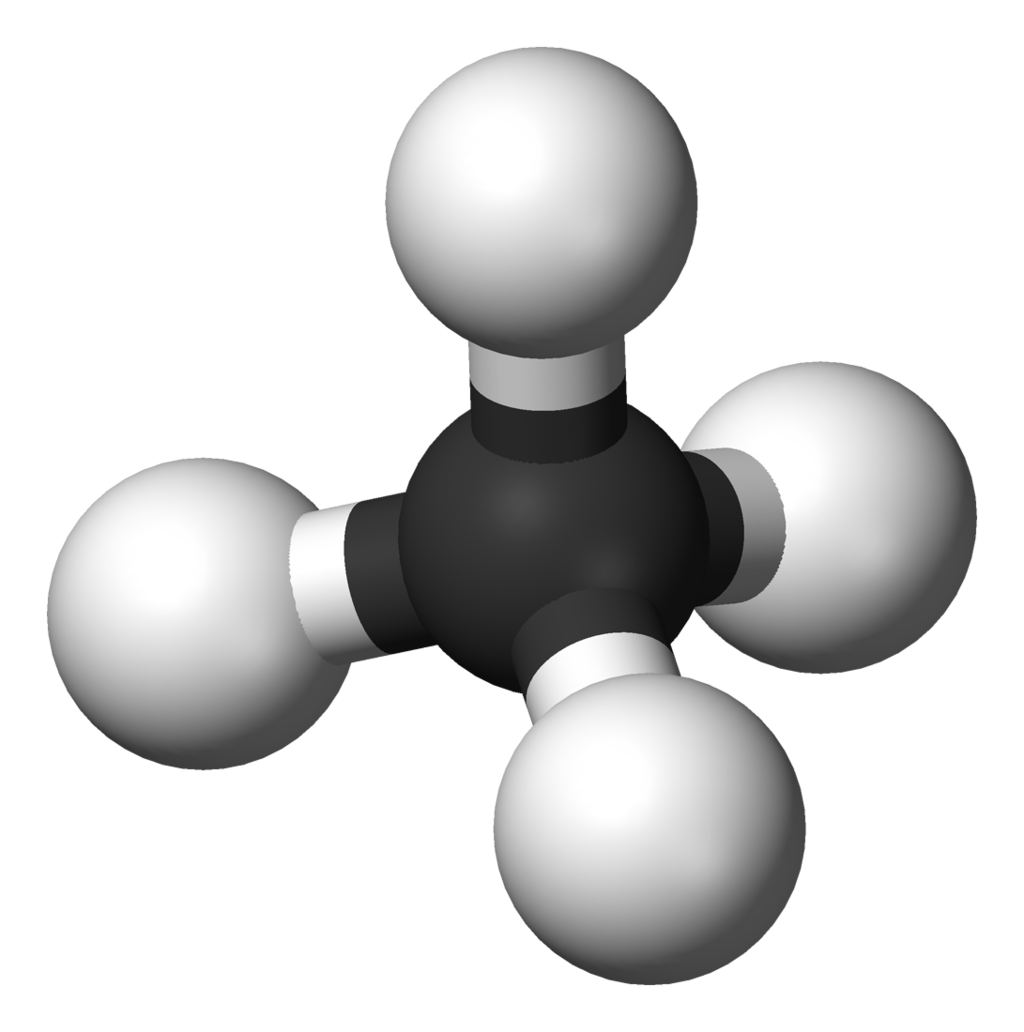 Ball-and-stick model of the methane molecule, CH4. Methane is part of a homologous series known as the alkanes, which contain single bonds only.
Ball-and-stick model of the methane molecule, CH4. Methane is part of a homologous series known as the alkanes, which contain single bonds only. |
|
|
|
hydrogen bond
A hydrogen bond (often informally abbreviated H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative atom or group, particularly the second-row elements nitrogen (N), oxygen (O), or fluorine (F)—the hydrogen bond donor (Dn)—and another electronegative atom bearing a lone pair of electrons—the hydrogen bond acceptor (Ac). Such an interacting system is generally denoted Dn–H···Ac, where the solid line denotes a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The use of three centered dots for the hydrogen bond is specifically recommended by the IUPAC. While hydrogen bonding has both covalent and electrostatic contributions, and the degrees to which they contribute are currently debated, the present evidence strongly implies that the primary contribution is covalent.
Hydrogen bonds can be intermolecular (occurring between separate molecules) or intramolecular (occurring among parts of the same molecule). Depending on the nature of the donor and acceptor atoms which constitute the bond, their geometry, and environment, the energy of a hydrogen bond can vary between 1 and 40 kcal/mol. This makes them somewhat stronger than a van der Waals interaction, and weaker than fully covalent or ionic bonds. This type of bond can occur in inorganic molecules such as water and in organic molecules like DNA and proteins. (W)
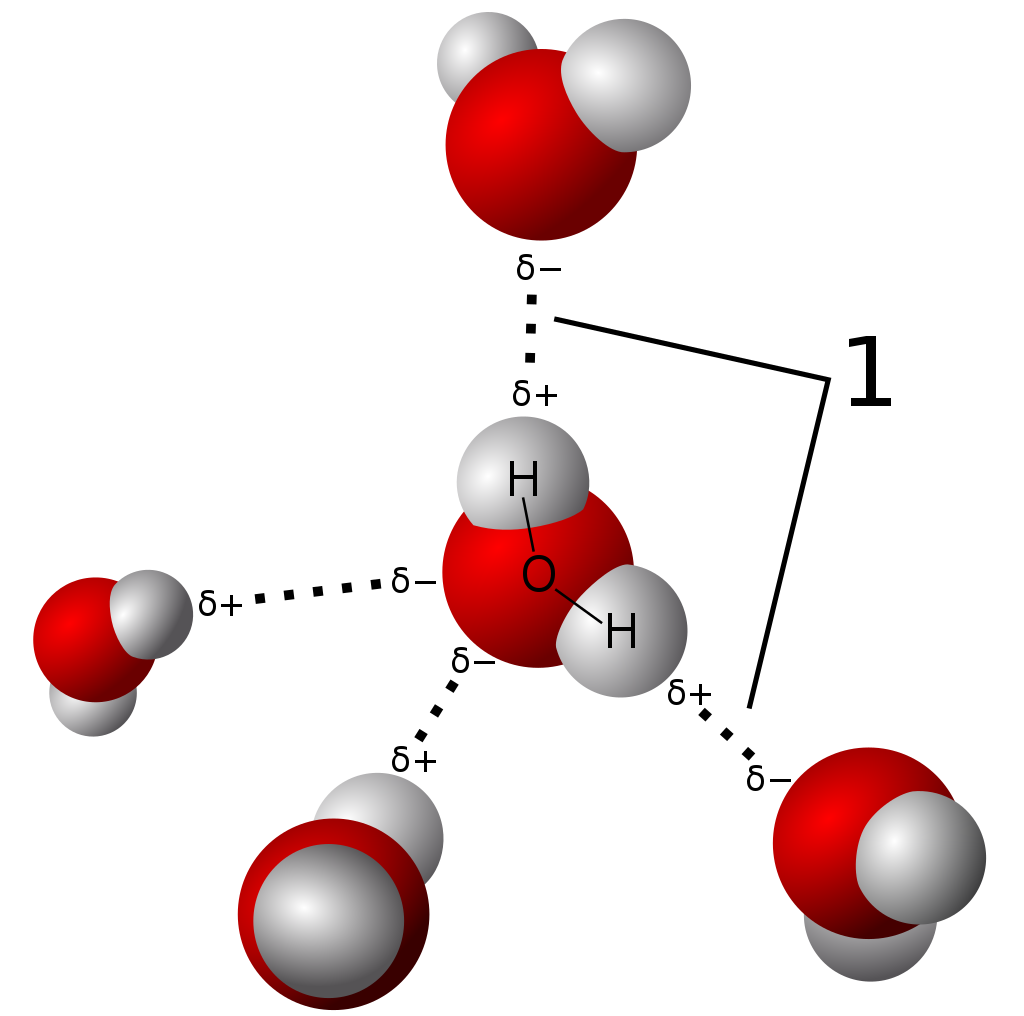
Model of hydrogen bonds (1) between molecules of water. |
|
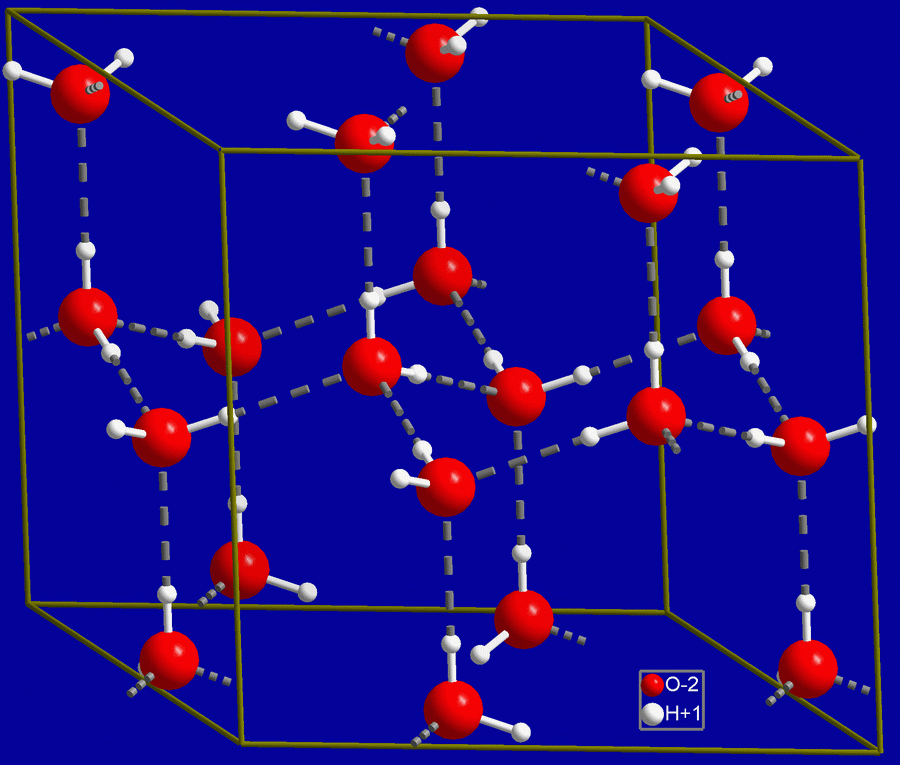
Crystal structure of hexagonal ice. Gray dashed lines indicate hydrogen bonds. |
|
|
|
hydrogen bonds in polymers
Hydrogen bonding plays an important role in determining the three-dimensional structures and the properties adopted by many synthetic and natural proteins. Compared to the C-C, C-O, and C-N bonds that comprise most polymers, hydrogen bonds are far weaker, perhaps 5%. Thus, hydrogen bonds can be broken by chemical or mechanical means while retaining the basic structure of the polymer backbone. This hierarchy of bond strengths (covalent bonds being stronger than hydrogen-bonds being stronger than van der Waals forces) is key to understanding the properties of many materials. (W)
|
|
hydrolysis
Hydrolysis (from Ancient Greek hydro- 'water', and lysis 'to unbind') is any chemical reaction in which a molecule of water ruptures one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile.
Biological hydrolysis is the cleavage of biomolecules where a water molecule is consumed to effect the separation of a larger molecule into component parts. When a carbohydrate is broken into its component sugar molecules by hydrolysis (e.g., sucrose being broken down into glucose and fructose), this is recognized as saccharification.
Hydrolysis reactions can be the reverse of a condensation reaction in which two molecules join together into a larger one and eject a water molecule. Thus hydrolysis adds water to break down, whereas condensation builds up by removing water and any other solvents. Some hydration reactions are hydrolyses. (W)
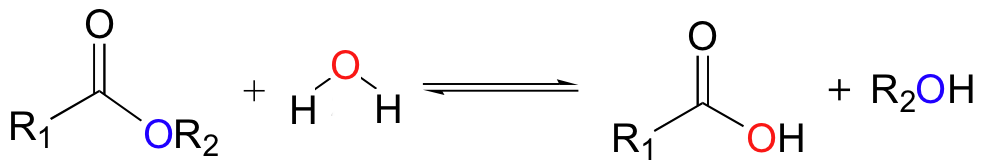
Generic mechanism for a hydrolysis reaction. (The 2-way yield symbol indicates an equilibrium in which hydrolysis and condensation can go both ways.). |
|
|
|
hydroxy group
A hydroxy or hydroxyl group is the entity with the formula OH. It contains oxygen bonded to hydrogen. In organic chemistry, alcohols and carboxylic acids contain hydroxy groups. Both the negatively charged anion OH−, called hydroxide, and the neutral radical ·OH, known as the hydroxyl radical, consist of an unbounded hydroxyl group.
According to IUPAC rules, the term hydroxyl refers to the hydroxyl radical (·OH) only, while the functional group −OH is called hydroxy group. (W)
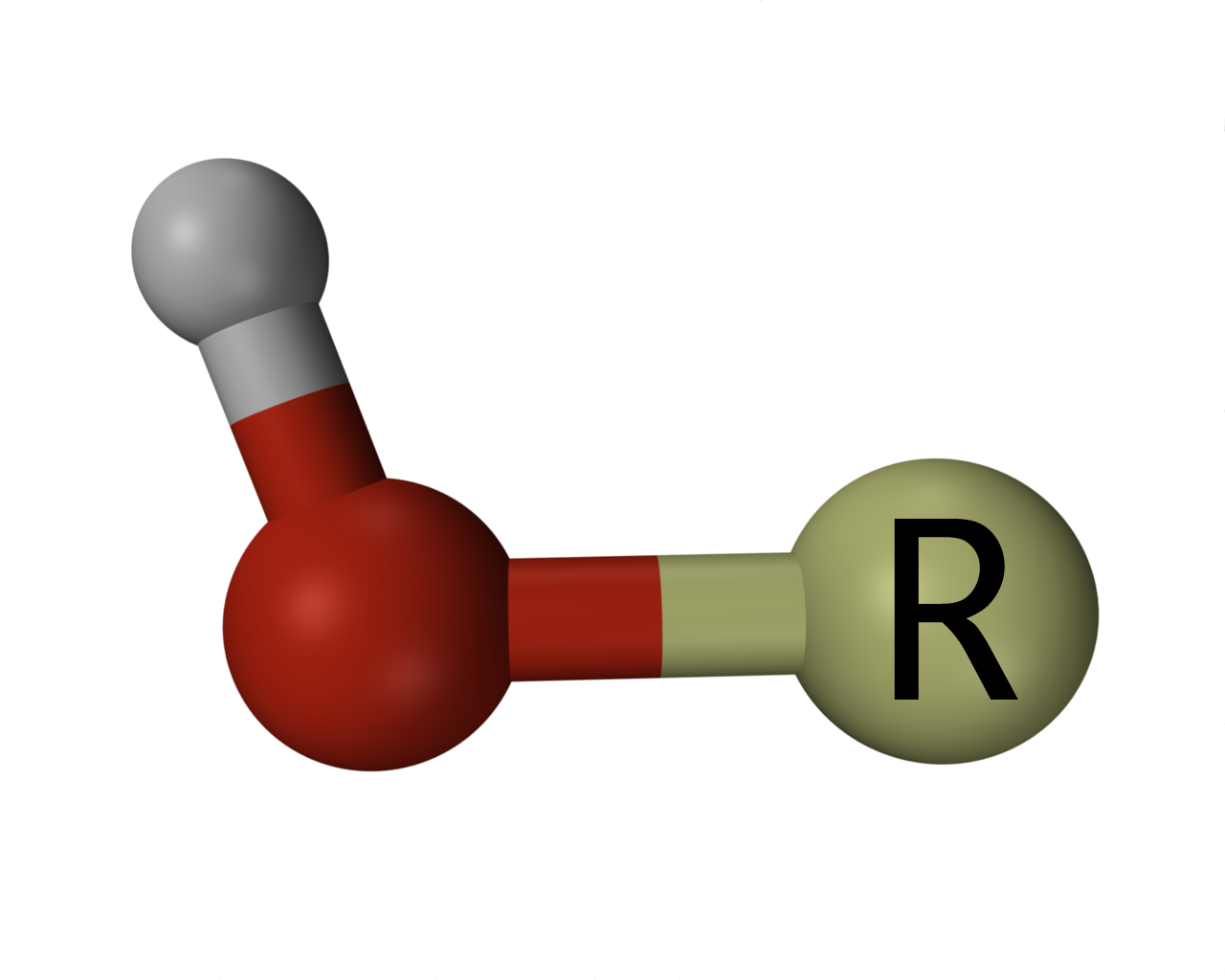 Representation of an organic hydroxy group, where R represents a hydrocarbon or other organic moiety, the red and grey spheres represent oxygen and hydrogen atoms respectively, and the rod-like connections between these, covalent chemical bonds.
Representation of an organic hydroxy group, where R represents a hydrocarbon or other organic moiety, the red and grey spheres represent oxygen and hydrogen atoms respectively, and the rod-like connections between these, covalent chemical bonds. |
|
| |
|
|
|
hydroxyl radical
The hydroxyl radical, •OH, is the neutral form of the hydroxide ion (OH−). Hydroxyl radicals are highly reactive (easily becoming hydroxy groups) and consequently short-lived; however, they form an important part of radical chemistry. Most notably hydroxyl radicals are produced from the decomposition of hydroperoxides (ROOH) or, in atmospheric chemistry, by the reaction of excited atomic oxygen with water. It is also an important radical formed in radiation chemistry, since it leads to the formation of hydrogen peroxide and oxygen, which can enhance corrosion and SCC in coolant systems subjected to radioactive environments. Hydroxyl radicals are also produced during UV-light dissociation of H2O2 (suggested in 1879) and likely in Fenton chemistry, where trace amounts of reduced transition metals catalyze peroxide-mediated oxidations of organic compounds. (W)
 Stick model of the hydroxyl radical with molecular orbitals.
Stick model of the hydroxyl radical with molecular orbitals. |
|
| |
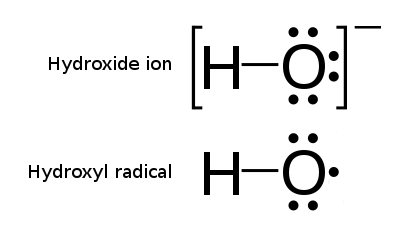 Comparison of a hydroxide ion and a hydroxyl radical.
Comparison of a hydroxide ion and a hydroxyl radical. |
|
📂Biological significance
Biological significance (W)
|
|
|
|